This work was support by the Yale Cooperative Center of Excellence in Hematology (NIH/NIDDK U54 DK106857).
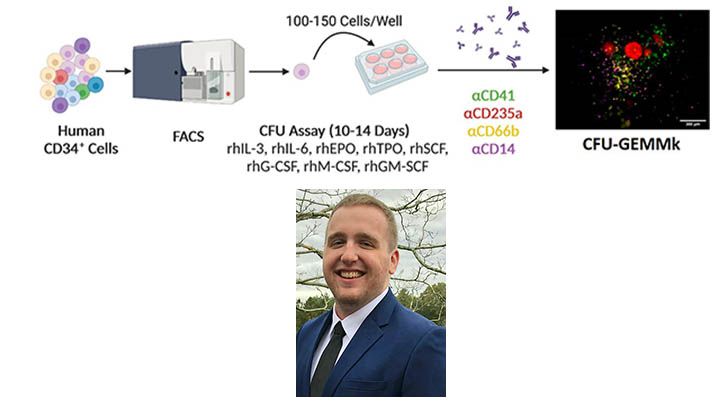
Colony forming unit (CFU) assays are a powerful tool in hematopoietic research as they allow researchers to functionally test the lineage potential of individual stem and progenitor cells. Assaying for lineage potential is important for determining and validating the identity of progenitor populations isolated by methods such as fluorescence activated cell sorting (FACS). However, current methods for CFU assays are limited in their ability to robustly assay multipotent progenitors with the ability to differentiate down the myeloid, erythroid and megakaryocytic lineages due to a deficiency of certain growth factors necessary for certain lineage outputs. In addition, manual counting of colony types is subjective resulting in variability of user-based assessments of cell types based on colony and cell morphologies. We demonstrate that addition of G-CSF, M-CSF, and GM-CSF into collagen-based MegaCult medium containing IL-3, IL-6, SCF, EPO, and TPO allows for the differentiation of common myeloid progenitors (CMP) into expected proportions of colonies containing granulocytic (G), monocytic (M), erythroid (E), and megakaryocytic (Mk) cells. Additionally, we demonstrate a high throughput objective method utilizing in situ immunofluorescence (IF) with anti-CD66b, anti-CD14, anti-CD235a, and anti-CD41 to detect G, M, E, and Mk cells, respectively. Thus, our improvements to the culture conditions and method for assay readout increase the accuracy, reproducibility, and throughput of the myeloid CFU assay.
