New “discrete state” model of hematopoiesis published in Nature Immunology. We propose a hierarchical model of hematopoiesis where stable “discrete states” serve as key regulatory nodes. @KyleFerchen figured out how to integrate CITE-seq, TEA-seq and InfinityFlow to link cell surface markers, gene expression, and chromatin accessibility to underlying gene regulatory networks (GRNs). These links facilitated physical isolation of genomic-cluster-defined populations to test their developmental potentials.
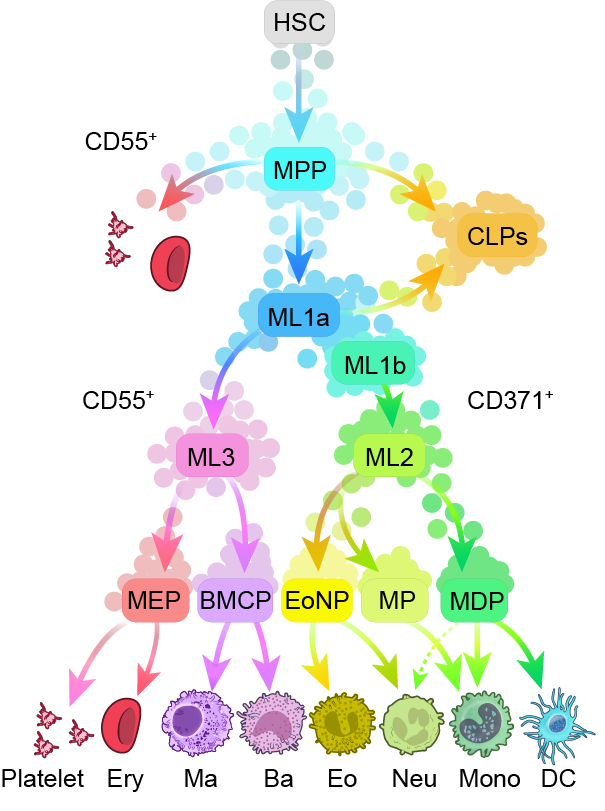
Large-scale, unbiased single-cell genomics studies of complex developmental compartments, such as hematopoiesis, have inferred novel cell states and trajectories; however, further characterization has been hampered by difficulty isolating cells corresponding to discrete genomic states. To address this, we present a framework that integrates multimodal single-cell analyses (RNA, surface protein and chromatin) with high-dimensional flow cytometry and enables semiautomated enrichment and functional characterization of diverse cell states. Our approach combines transcription factor expression with chromatin activity to uncover hierarchical gene regulatory networks driving these states. We delineated and isolated rare bone marrow Lin−Sca−CD117+CD27+ multilineage cell states (‘MultiLin’), validated predicted lineage trajectories and mapped differentiation potentials. Additionally, we used transcription factor activity on chromatin to trace and isolate multilineage progenitors undergoing multipotent to oligopotent lineage restriction. In the proposed model of steady-state hematopoiesis, discrete states governed developmental trajectories. This framework provides a scalable solution for isolating and characterizing novel cell states across different biological systems.

